MAKE A MEME
View Large Image
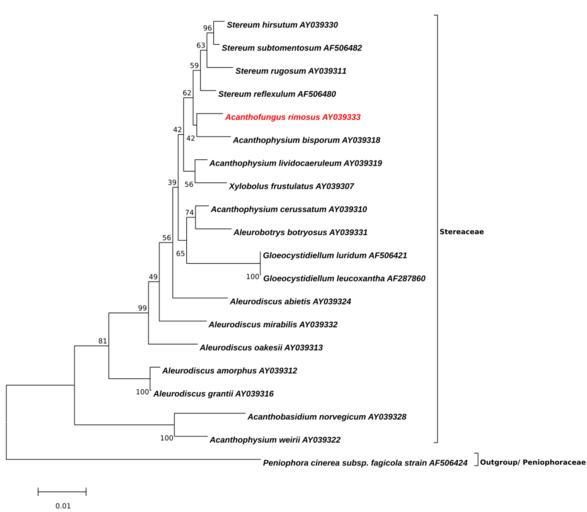
| View Original: | Acanthofungus-Minimum Evolution-Tree.svg (154x134) | |||
| Download: | Original | Medium | Small | Thumb |
| Courtesy of: | commons.wikimedia.org | More Like This | ||
| Keywords: Acanthofungus-Minimum Evolution-Tree.svg Figure Evolutionary relationships of Acanthofungus rimosus and related taxa <br/> The evolutionary history was inferred using the Minimum Evolution method 1 The optimal tree with the sum of branch length 0 24677858 is shown The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test 1000 replicates are shown next to the branches 2 The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree The evolutionary distances were computed using the Kimura 2-parameter method 3 and are in the units of the number of base substitutions per site The ME tree was searched using the Close-Neighbor-Interchange CNI algorithm 4 at a search level of 2 The Neighbor-joining algorithm 5 was used to generate the initial tree The analysis involved 20 nucleotide sequences All positions with less than 95 site coverage were eliminated That is fewer than 5 alignment gaps missing data and ambiguous bases were allowed at any position There were a total of 891 positions in the final dataset Evolutionary analyses were conducted in http //megasoftware net/ MEGA6 6 <br/> 1 Rzhetsky A and Nei M 1992 A simple method for estimating and testing minimum evolution trees Molecular Biology and Evolution 9 945-967 <br/> 2 Felsenstein J 1985 Confidence limits on phylogenies An approach using the bootstrap Evolution 39 783-791 <br/> 3 Kimura M 1980 A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences Journal of Molecular Evolution 16 111-120 <br/> 4 Nei M and Kumar S 2000 Molecular Evolution and Phylogenetics Oxford University Press New York <br/> 5 Saitou N and Nei M 1987 The neighbor-joining method A new method for reconstructing phylogenetic trees Molecular Biology and Evolution 4 406-425 <br/> 6 Tamura K Stecher G Peterson D Filipski A and Kumar S 2013 MEGA6 Molecular Evolutionary Genetics Analysis version 6 0 Molecular Biology and Evolution 30 2725-2729 <br/> ; List with GenBank Sequences which were used for the ML-Tree http //www ncbi nlm nih gov/nuccore/AY039328 Acanthobasidium norvegicum AY039328 http //www ncbi nlm nih gov/nuccore/AY039333 Acanthofungus rimosus AY039333 http //www ncbi nlm nih gov/nuccore/AY039318 Acanthophysium bisporum AY039318 http //www ncbi nlm nih gov/nuccore/AY039310 Acanthophysium cerussatum AY039310 http //www ncbi nlm nih gov/nuccore/AY039319 Acanthophysium lividocaeruleum AY039319 http //www ncbi nlm nih gov/nuccore/AY039322 Acanthophysium weirii AY039322 http //www ncbi nlm nih gov/nuccore/AY039331 Aleurobotrys botryosus AY039331 http //www ncbi nlm nih gov/nuccore/AY039324 Aleurodiscus abietis AY039324 http //www ncbi nlm nih gov/nuccore/AY039312 Aleurodiscus amorphus AY039312 http //www ncbi nlm nih gov/nuccore/AY039316 Aleurodiscus grantii AY039316 http //www ncbi nlm nih gov/nuccore/AY039332 Aleurodiscus mirabilis AY039332 http //www ncbi nlm nih gov/nuccore/AY039313 Aleurodiscus oakesii AY039313 http //www ncbi nlm nih gov/nuccore/AF287860 Gloeocystidiellum leucoxantha AF287860 http //www ncbi nlm nih gov/nuccore/AF506421 Gloeocystidiellum luridum AF506421 http //www ncbi nlm nih gov/nuccore/AF506424 Peniophora cinerea subsp fagicola strain AF506424 http //www ncbi nlm nih gov/nuccore/AY039330 Stereum hirsutum AY039330 http //www ncbi nlm nih gov/nuccore/AF506480 Stereum reflexulum AF506480 http //www ncbi nlm nih gov/nuccore/AY039311 Stereum rugosum AY039311 http //www ncbi nlm nih gov/nuccore/AF506482 Stereum subtomentosum AF506482 http //www ncbi nlm nih gov/nuccore/AY039307 Xylobolus frustulatus AY039307 own 2014-08-22 Thkgk Cc-zero Acanthofungus Acanthofungus rimosus | ||||